MrOlm/drep: Rapid comparison and dereplication of genomes
推荐用conda安装
conda create -n drep
conda activate drep
conda install drep -c bioconda也可以用pip,但是有一些依赖的包可能需要自己下
pip install drep
安装版本V3.2.2
drep:微生物基因组快速去冗余-文章解读+帮助文档+实战教程

1. dRep需要依赖一些软件
运行
$ dRep check_dependenciesmash.................................... !!! ERROR !!! (location = None)
nucmer.................................. !!! ERROR !!! (location = None)
checkm.................................. all good (location =
ANIcalculator........................... !!! ERROR !!! (location = None)
prodigal................................ all good (location = /usr/bin/prodigal)
centrifuge.............................. !!! ERROR !!! (location = None)
nsimscan................................ !!! ERROR !!! (location = None)
fastANI................................. !!! ERROR !!! (location = None)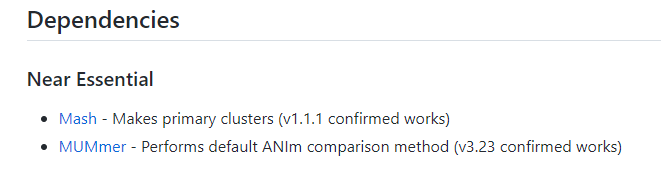
这两个是必须的
可以单独安装,也可以让conda安装
这两个应该都行
conda install -c bioconda mash
conda install -c bioconda/label/cf201901 mashconda install -c bioconda mummer
conda install -c bioconda/label/cf201901 mummermash的安装
Mash: fast genome and metagenome distance estimation using MinHash | Genome Biology | Full Text
marbl/Mash: Fast genome and metagenome distance estimation using MinHash
Release Mash v2.3 · marbl/Mash
下载之后,安装就ok了
nucmer的安装
mummer4/mummer: Mummer alignment tool
mummer/INSTALL.md at master · mummer4/mummer
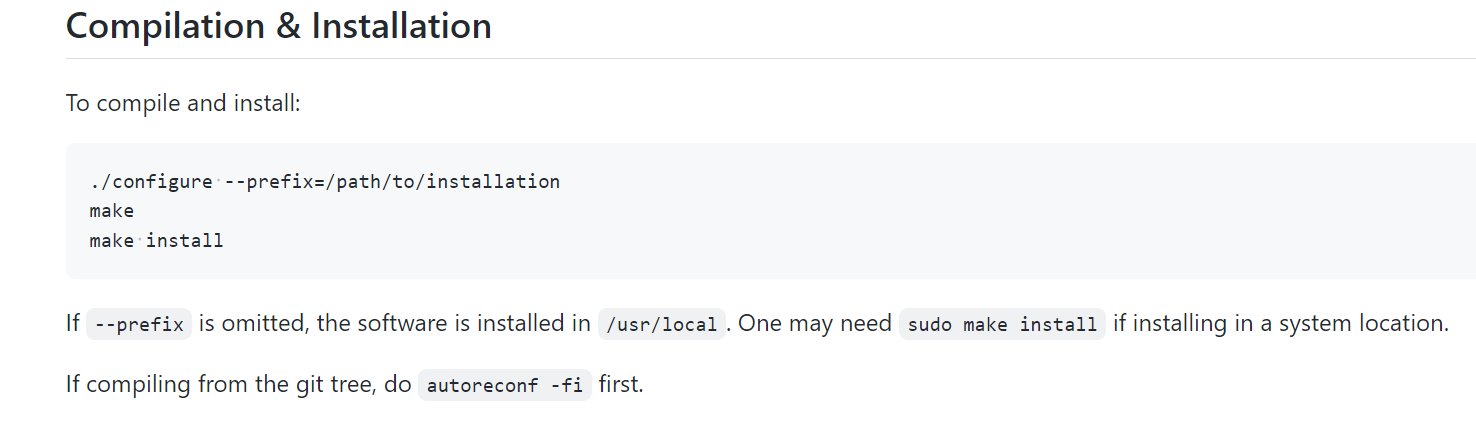
然后
这些是可选的
我下centrifuge的时候,发现我的版本可能高了,不适配了
不用都下,用不到就先不下,报错了再下也不迟
2. 实战
Try1
##模拟数据来源刘永鑫
(drep) chenl 16:32:14 ~/drep_try/fa
$ ls
B4018L.2.fa K4093L.5.fa K4096L.2.fa L4105L.2.fa W4194L.3.fa W4194L.6.fa
$ dRep dereplicate out1 -g ./fa/*.fa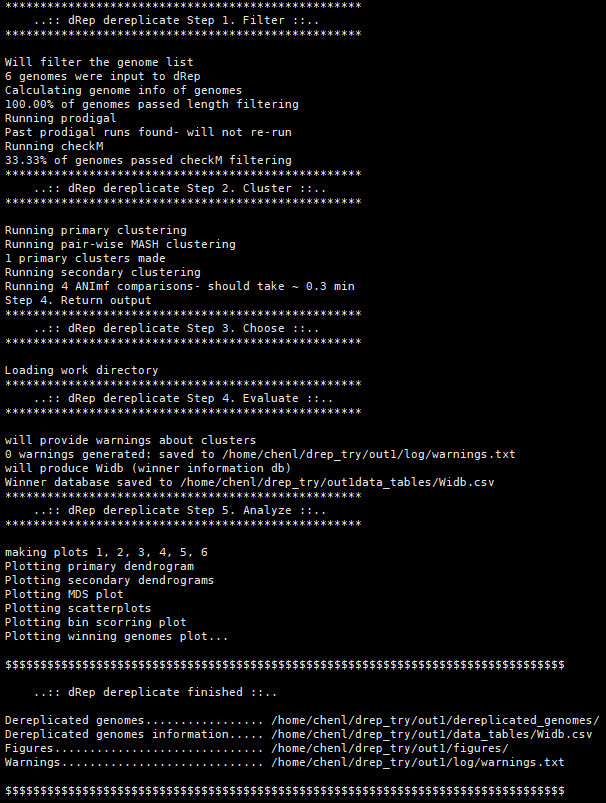
checkm的时间比较久,然后啪叽就成功了
Succeed:happy:
Try2
dRep dereplicate ./ -g bin/*.fa -sa 0.95 -nc 0.30 -p 24 -comp 50 -con 10 -sa S_ANI, --S_ani S_ANI
二级聚类为99% ANI threshold to form secondary clusters (default:
0.99)
-nc COV_THRESH, --cov_thresh COV_THRESH
最小的重叠是10% Minmum level of overlap between genomes when doing
secondary comparisons (default: 0.1)
- p 线程
- comp 完整度
- 污染度
Try3
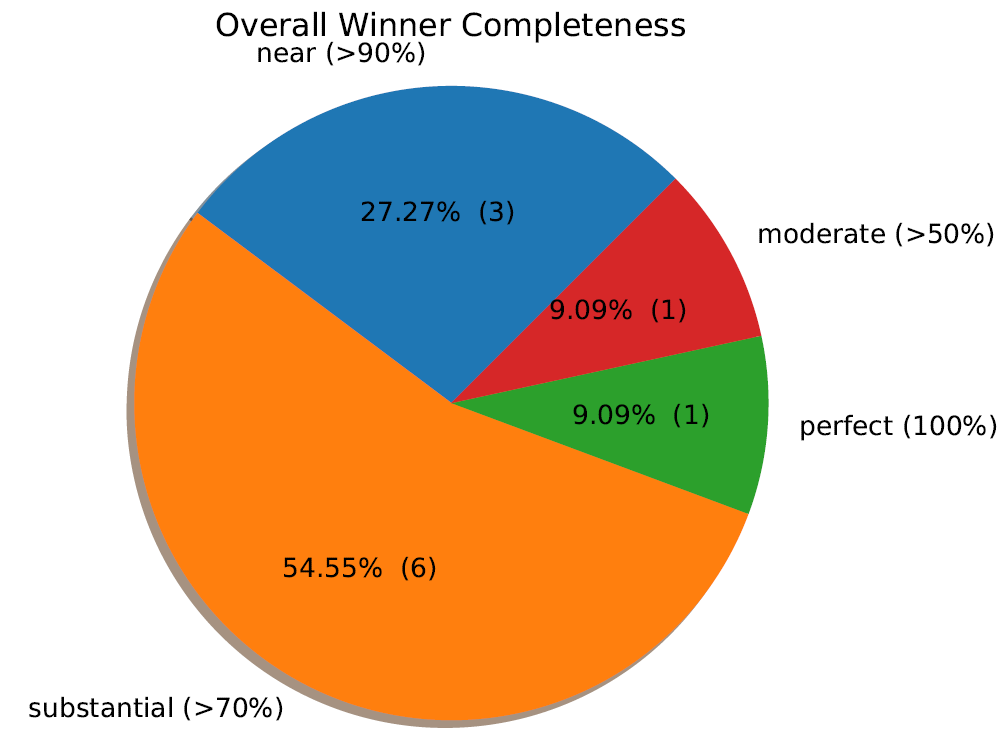
3. 结果
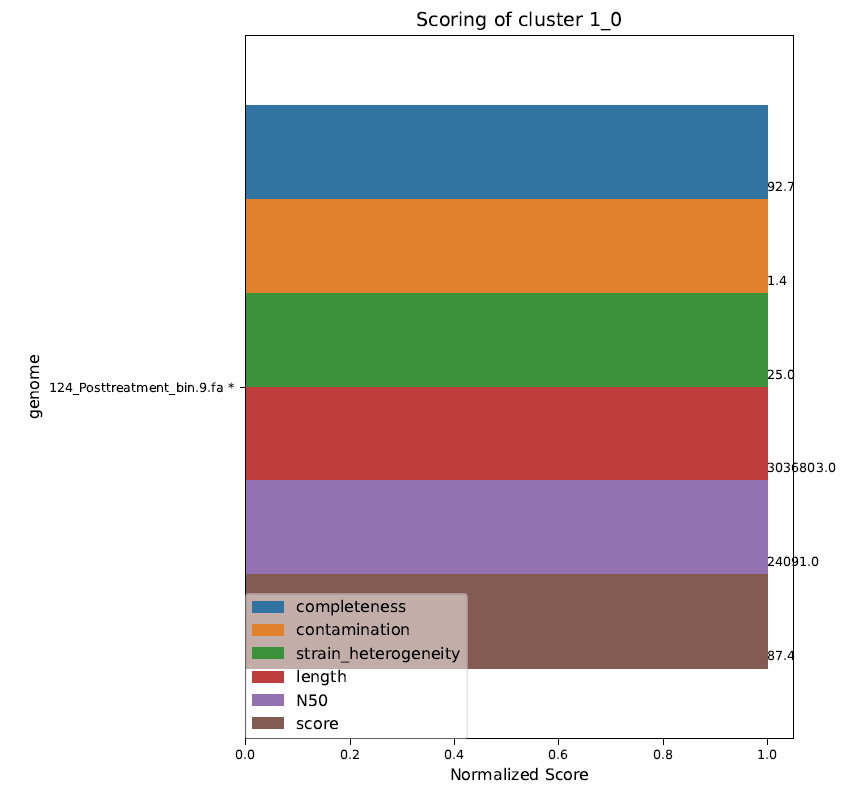
Cluster_scoring
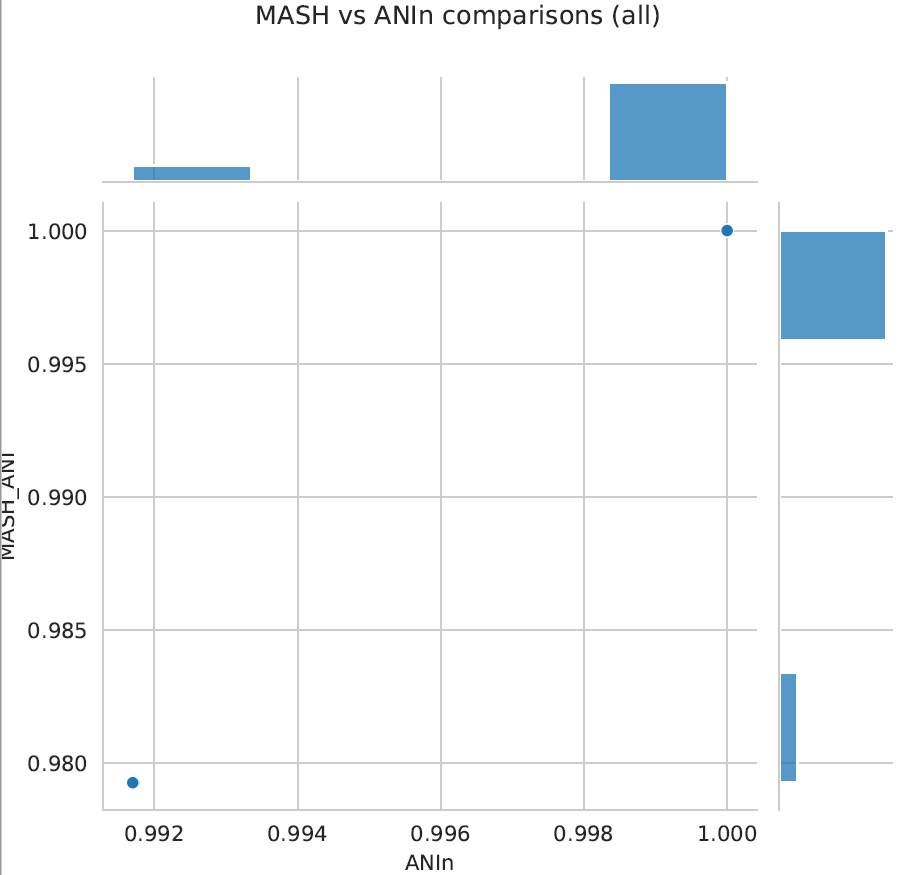
Clustering_scatterplots
Primary_clustering_dendrogram
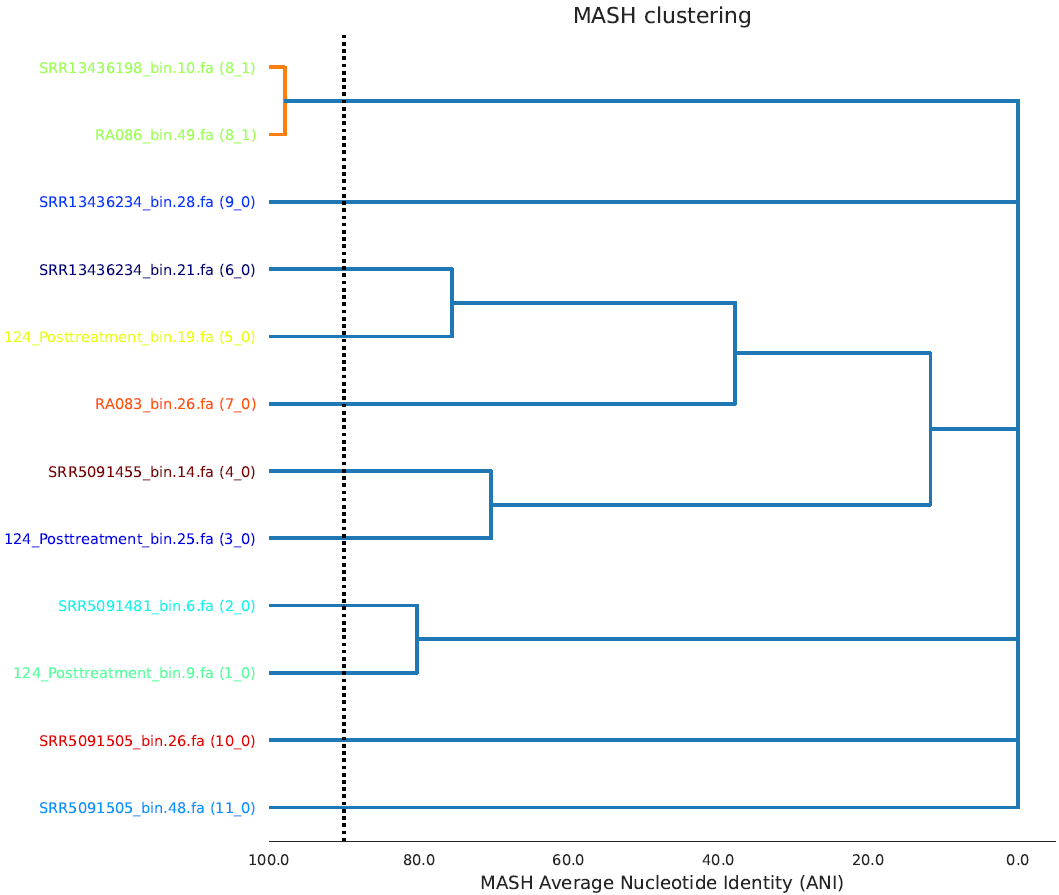
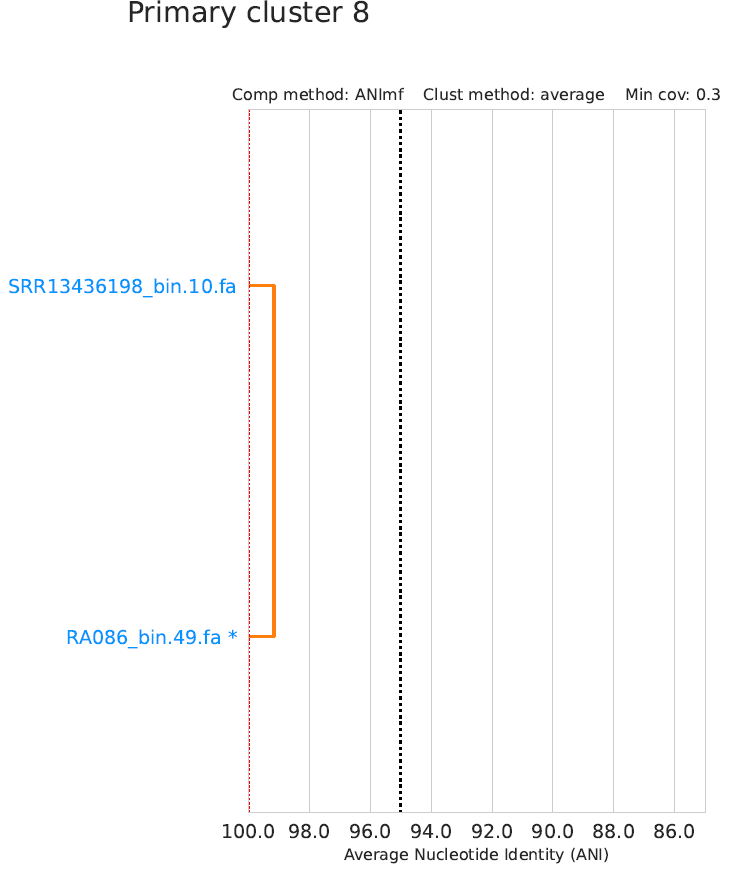
Secondary_clustering_dendrograms
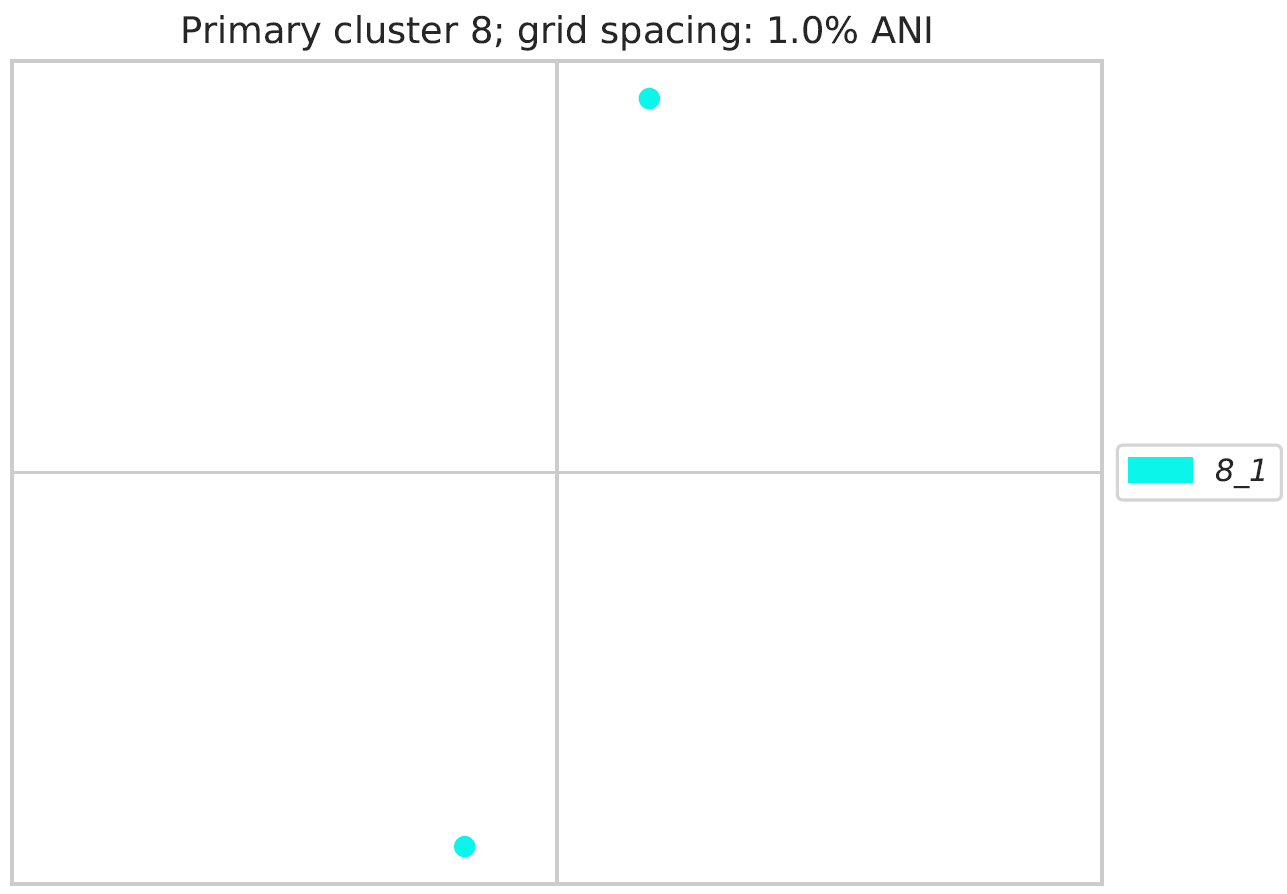
Secondary_clustering_MDS
Winning_genomes